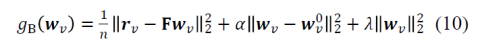[I7R8]
Realistic models of the primate visual system have many millions of parameters. A vision model needs substantial capacity to store the required knowledge about what things look like. Brain activity data are costly, so typically do not suffice to set the parameters of these models. Recent progress has benefited from direct learning of the required knowledge from category-labeled image sets. Nevertheless further fitting with brain-activity data is required to learn about the relative prevalence of the different computational features (and of linear combinations of the features) in each cortical area and to accurately predict representations of novel images (not used in setting model parameters).
Each individual brain is unique. A key challenge is to hold on to what we’ve learned by fitting a visual encoding model to one subject exposed to one set of images when we move on to new experiments. Traditionally, we make inferences about the computational mechanisms with a given data set and hold on to those abstract insights, e.g. that model ResNet beats model AlexNet at predicting ventral visual responses. Ideally, we would be able to hold on to more detailed parametric information learned on one data set as we move on to other data sets.
Wen, Shi, Chen & Liu (pp2017) develop a Bayesian approach to learning encoding models (linear combinations of the features of deep neural networks) incrementally across subjects and stimulus sets. The initial model is fitted with a 0-mean prior on the weights (L2 penalty). The resulting encoding model for each fMRI voxel has a Gaussian posterior over the weights for each feature of the deep net model. The Gaussian posterior is assumed to be isotropic, avoiding the need for a separate variance parameter for each feature (let alone a full covariance matrix).
The results are compelling. Using the posteriors inferred from previous subjects as priors for new subjects substantially increases a model’s prediction performance. This is consistent with the observation that models generalize quite well to new subjects, even without subject-specific fitting. Importantly, the transfer of the weight knowledge from one subject to the next works even when using different stimulus sets in different subjects.
This work takes a first step in the direction of the exciting possibility of incremental learning of complex models across hundreds or thousands of subjects and millions of stimuli (acquired in labs around the world).
It is interesting to consider the implementation of the inference procedure. Although Bayesian in motivation, the implementation uses L2 penalities for deviation of the weights wv from the previous weights estimate wv0 and from zero. The respective penalty factors α and λ are determined by crossvalidation so as to best predict the new data. This procedure makes a lot of sense. However, it is a bit at a tension with a pure Bayesian approach in two ways: (1) In a pure Bayesian approach, the previous data set should determine the width of the posterior, which becomes the prior for the next data set. Here the width of the prior is adjusted (via α) to optimize prediction performance. (2) In a pure Bayesian approach, the 0-mean prior would be absorbed into the first model’s posterior and would not enter into into the inference again with every update of the posterior with new data.
The cost function for predicting the response profile vector rv (# stimuli by 1) for fMRI voxel v from deep net feature responses F (# stimuli by # features) is:
While the crossvalidation procedure makes sense for optimizing prediction accuracy on the present data set, I wonder if it is optimal in the bigger picture of integrating the knowledge across many studies. The present data set will reflect only a small portion of stimulus space and one subject, so should not get to downweight a prior based on much more comprehensive data.
Strengths
- Addresses an important challenge and suggests exciting potential for big-data learning of computational models across studies and labs.
- Presents a straightforward and well-motivated method for incremental learning of encoding model weights across studies with different subjects and different stimuli.
- Results are compelling: Using the prior information helps the performance of an encoding model a lot when the training data for the new subject is limited.
Weaknesses
- The posterior over the weights vector is modeled as isotropic. It would be good to allow different degrees of certainty for different features and, better yet, to model the dependencies between the weights of different features. (However, such richer models might be challenging to estimate in practice.)
- The prior knowledge transferred from previous studies consists only in the MAP estimate of the weight vector for each voxel.
- The method assumes that a precise intersubject spatial-correspondence mapping is given. Such mappings might not exist and are costly to approximate with functional data.
Suggestions for improvement
(1) Explore and/or discuss if a prior with feature-specific variance might be feasible. Explore whether inferring a posterior distribution over weights using a mean weight vector and feature-specific variances brings even better results. I guess this is hard when there are millions of features.
(2) Consider dropping the assumption that a precise correspondence mapping is given and infer a multinormal posterior over local weight vectors. The model assumes that we have a precise intersubject spatial-correspondence mapping (from cortical alignment based either on anatomical or functional data). It seems more versatile and statistically preferable not to rely on a precise (i.e. voxel-to-voxel) correspondence mapping, but to simultaneously address the correspondence and incremental weight-learning problem. We could assume that an imprecise correspondence mapping is given. For corresponding brain locations in the previous and current subject (subjects 1 and 2), subject-1 encoding models within a small spherical region around the target location could be used to define a prior for fitting an encoding model to the target voxel for subject 2. Such a prior should be a probability distribution over weight vectors, which could be characterized by the second moment of the weight vector distribution. Regularization, such as optimal shrinkage to a diagonal target or (when there are too many features) simply the assumption that the second moment is diagonal could be used to make this approach feasible. In either case, the goal would be to pool the posterior distributions across voxels within the small sphere and summarize the resulting distribution (e.g. as a multinormal). I realize that this might be beyond the scope of the current study. It is not a requirement for this paper.
(3) Clarify the terminology used for the estimation procedures. What is referred to as “maximum likelihood estimation” uses an L2 penalty on the weights, amounting to Bayesian inference of the weights with a 0-mean Gaussian prior. This is not a maximum likelihood estimator. Please correct this (or explain in case I am mistaken).
(4) Consider how to ensure that the prior has an appropriate width (and the prior evidence thus appropriate weight). Should a more purely Bayesian approach be taken, where the width of the posterior is explicitly inferred and becomes the width of the prior? Should the crossvalidation setting of the hyperparameters use a very varied test set to prevent the current (possibly narrowly specialized) data set from being given too much weight? Should the amount of data contributing to the prior model and the amount of data in the present set (and optionally the noise level) be used to determine the relative weighting?