Open review of “Post-Publication Peer Review for Real”
by Koki Ikeda, Yuki Yamada, and Kohske Takahashi (pp2020)
[I7R8]
Our system of pre-publication peer review is a relict of the age when the only way to disseminate scientific papers was through print media. Back then the peer evaluation of a new scientific paper had to precede its publication, because printing (on actual paper if you can believe it) and distributing the physical print copies is expensive. Only a small selection of papers could be made accessible to the entire community.
Now the web enables us to make any paper instantly accessible to the community at negligible cost. However, we’re still largely stuck with pre-publication peer review, despite its inherent limitations: to a small number of preselected reviewers who operate in isolation from and without the scrutiny of the community.
People familiar with the web who have considered, from first principles, how a scientific peer review system should be designed tend to agree that it’s better to make a new paper publicly available first, so the community can take note of the work and a broader set of opinions can contribute to the evaluation. Post-publication peer review also enables us to make the evaluation transparent: Peer reviews can be open responses to a new paper. Transparency promises to improve reviewers’ motivation to be objective, especially if they choose to sign and take responsibility for their reviews.
We’re still using the language of a bygone age, whose connotations make it hard to see the future clearly:
- A paper today is no longer made of paper — but let’s stick with this one.
- A preprint is not something that necessarily precedes the publication in print media. A better term would be “published paper”.
- The term publication is often used to refer to a journal publication. However, preprints now constitute the primary publications. First, a preprint is published in the real sense: the sense of having been made publicly available. This is in contrast to a paper in Nature, say, which is locked behind a paywall, and thus not quite actually published. Second, the preprint is the primary publication in that it precedes the later appearance of the paper in a journal.
Scientists are now free to use the arXiv and other repositories (including bioRxiv and PsyArXiv) to publish papers instantly. In the near future, peer review could be an open and open-ended process. Of course papers could still be revised and might then need to be re-evaluated. Depending on the course of the peer evaluation process, a paper might become more visible within its field, and perhaps even to a broader community. One way this could happen is through its appearance in a journal.
The idea of post-publication peer review has been around for decades. Visions for open post-publication peer review have been published. Journals and conferences have experimented with variants of open and post-publication peer review. However, the idea has yet to revolutionize the scientific publication system.
In their new paper entitled “Post-publication Peer Review for Real”, Ikeda, Yamada, and Takahashi (pp2020) argue that the lack of progress with post-publication peer review reflects a lack of motivation among scientists to participate. They then present a proposal to incentivize post-publication peer review by making reviews citable publications published in a journal. Their proposal has the following features:
- Any scientist can submit a peer review on any paper within the scope of the journal that publishes the peer reviews (the target paper could be published either as a preprint or in any journal).
- Peer reviews undergo editorial oversight to ensure they conform to some basic requirements.
- All reviews for a target paper are published together in an appealing and readable format.
- Each review is a citable publication with a digital object identifier (DOI). This provides a new incentive to contribute as a peer reviewer.
- The reviews are to be published as a new section of an existing “journal with high transparency”.
Ikeda at al.’s key point that peer reviews should be citable publications is solid. This is important both to provide an incentive to contribute and also to properly integrate peer reviews into the crystallized record of science. Making peer reviews citable publications would be a transformative and potentially revolutionary step.
The authors are inspired by the model of Behavioral and Brain Sciences (BBS), an important journal that publishes theoretical and integrative perspective and review papers as target articles, together with open peer commentary. The “open” commentary in BBS is very successful, in part because it is quite carefully curated by editors (at the cost of making it arguably less than entirely “open” by modern standards).
BBS was founded by Stevan Harnad, an early visionary and reformer of scientific publishing and peer review. Harnad remained editor-in-chief of BBS until 2002. He explored in his writings what he called “scholarly skywriting“, imagining a scientific publication system that combines elements of what is now known as open-notebook science and research blogging with preprints, novel forms of peer review, and post-publication peer commentary.
If I remember correctly, Harnad drew a bold line between peer review (a pre-publication activity intended to help authors improve and editors select papers) and peer commentary (a post-publication activity intended to evaluate the overall perspective or conclusion of a paper in the context of the literature).
I am with Ikeda et al. in believing that the lines between peer review and peer commentary ought to be blurred. Once we accept that peer review must be post-publication and part of a process of community evaluation of new papers, the prepublication stage of peer review falls away. A peer review, then, becomes a letter to both the community and to the authors and can serve any combination of a broader set of functions:
- to explain the paper to a broader audience or to an audience in an adjacent field,
- to critique the paper at the technical and conceptual level and possibly question its conclusions,
- to relate it to the literature,
- to discuss its implications,
- to help the authors improve the paper in revision by adding experiments or analyses and improving the exposition of the argument in text and figures.
An example of this new form is the peer review you are reading now. I review only papers that have preprints and publish my peer reviews on this blog. This review is intended for both the authors and the community. The authors’ public posting of a preprint indicates that they are ready for a public response.
Admittedly, there is a tension between explaining the key points of the paper (which is essential for the community, but not for the authors) and giving specific feedback on particular aspects of the writing and figures (which can help the authors improve the paper, but may not be of interest to the broader community). However, it is easy to relegate detailed suggestions to the final section, which anyone looking only to understand the big picture can choose to skip.
Importantly, the reviewer’s judgment of the argument presented and how the paper relates to the literature is of central interest to both the authors and the community. Detailed technical criticism may not be of interest to every member of the community, but is critical to the evaluation of the claims of a paper. It should be public to provide transparency and will be scrutinized by some in the community if the paper gains high visibility.
A deeper point is that a peer review should speak to the community and to the authors in the same voice: in a constructive and critical voice that attempts to make sense of the argument and to understand its implications and limitations. There is something right, then, about merging peer review and peer commentary.
While reading Ikeda et al.’s review of the evidence that scientists lack motivation to engage in post-publication peer review, I asked myself what motivates me to do it. Open peer review enables me to:
- more deeply engage the papers I review and connect them to my own ideas and to the literature,
- more broadly explore the implications of the papers I review and start bigger conversations in the community about important topics I care about,
- have more legitimate power (the power of a compelling argument publicly presented in response to the claims publicly presented in a published paper),
- have less illegitimate power (the power of anonymous judgment in a secretive process that decides about publication of someone else’s work)
- take responsiblity for my critical judgments by subjecting them to public scrutiny
- make progress with my own process of scientific insight
- help envision a new form of peer review that could prove positively transformative
In sum, open post-publication peer review, to me, is an inherently more meaningful activity than closed pre-publication peer review. I think there is plenty of motivation for open post-publication peer review, once people overcome their initial uneasiness about going transparent. A broader discussion of researcher motivations for contributing to open post-publication peer review is here.
That said, citability and DOIs are essential, and so are the collating and readability of the peer reviews of a target paper. I hope Ikeda et al. will pursue their idea of publishing open post-publication peer reviews in a journal. Gradually, and then suddenly, we’ll find our way toward a better system.
Suggestions for improvements
(1) The proposal raises some tricky questions that the authors might want to address:
- Which existing “journal with high transparency” should this be implemented in?
- Should it really be a section in an existing journal or a new journal (e.g. the “Journal of Peer Reviews in Psychology”)?
- Are the peer reviews published immediately as they come in, or in bulk once there is a critical mass?
- Are new reviews of a target paper to be added on an ongoing basis in perpetuity?
- How are the target papers to be selected? Should their status as preprints or journal publications make any difference?
- Why do we need to stick with the journal model? Couldn’t commentary sections on preprint servers solve the problem more efficiently — if they were reinvented to provide each review also as a separate PDF with beautiful and professional layout, along with figure and LaTeX support and, critically, citability and DOIs?
Consider addressing some of these questions to make the proposal more compelling. In particular, it seems attractive to find an efficient solution linked to preprint servers to cover large parts of the literature. Can the need for editorial work be minimized and the critical incentive provided through beautiful layout, citability, and DOIs?
(2) Cite and and discuss some of Stevan Harnad’s contributions. Some of the ideas in this edited collection of visions for post-publication peer review may also be relevant.
A recent large-scale survey reported that 98% of researchers who participated in the study agreed that the peer-review system was important (or extremely important) to ensure the quality and integrity of science. In addition, 78.8% answered that they were satisfied (or very satisfied) with the current review system (Publon, 2018) . It is probably true that peer-review has been playing a significant role to control the quality of academic papers (Armstrong, 1997) . The latter result, however, is rather perplexing, since it has been well known that sometimes articles could pass through the system without their flaws being revealed (Hopewell et al., 2014) , results could not be
reproduced reliably (e.g. Open Science Collaboration, 2015) , decisions were said to be no better than a dice roll (Lindsey, 1988; Neff & Olden, 2006) , and inter-reviewer agreement was estimated to be very low (Bornmann et al., 2010).
(3) Consider disentangling the important pieces of evidence in the above passage a little more. “Perplexing” seems the wrong word here: Peer review can be simultaneously the best way to evaluate papers and imperfect. It would be good to separate mere evidence that mistakes happen (which appears unavoidable), from the stronger criticism that peer review is no better than random evaluations. A bit more detail on the cited results suggesting it is no better than random would be useful. Is this really a credible conclusion? Does it require qualifications?
The low reliability across reviewers is especially disturbing and raises serious concerns about the effectiveness of the system, because we now have empirical data showing that inter-rater agreement and precision could be very high, and they robustly predict the replicability of previous studies, when the information about others’ predictions are shared among predictors (Botvinik-Nezer et al., 2020; Camerer et al., 2016, 2018; Dreber et al., 2015; Forsell et al., 2019) . Thus, the secretiveness of the current system could be the unintended culprit of its suboptimality.
(4) Consider revising the above passage. Inter-reviewer agreement is an important metric to consider. However, even zero correlation between reviewer’s ratings does not imply that the reviews are random. Reviewers may focus on different criteria. For example, if one reviewer judged primarily the statistical justification of the claims and another primarily the quality of the writing, the correlation between their ratings could be zero. However, the average rating would be a useful indicator of quality. Averaging ratings in this context does not serve merely to reduce the noise in the evaluations, it also serves to compromise between different weightings of the criteria of quality.
Interaction among reviewers that enables them to adjust their judgments can fundamentally enhance the review process. However, inter-rater agreement is an ambiguous measure, when the ratings are not independent.
However, BBS commentary is different from them in terms of that it employs an “open” system so that anyone can submit the commentary proposal at will (although some commenters are arbitrarily chosen by the editor). This characteristic makes BBS commentary much more similar to PPPR than other traditional publications.
(5) Consider revising. Although BBS commentaries are nothing like traditional papers (typically much briefer statements of perspective on a target paper) and are a form of post-publication evaluation, they are also very distinct in form and content. I think Stevan Harnad made this point somewhere.
Next and most importantly, the majority of researchers find no problem with their incentives to submit an article as a BBS commentary, because they will be considered by many researchers and institutes to be equivalent to a genuine publication and can be listed on one’s CV. Therefore, researchers have strong incentives to actively publish their reviews on BBS .
(6) Consider revising. It’s a citable publication, yes. However, it’s in a minor category, nowhere near a primary research paper or a full review or perspective paper.
There seem to be at least two reasons for this uniqueness. Firstly, BBS is undoubtedly one of the most prestigious journals in psychology and its
related areas, with a 17.194 impact factor for the year 2018. Secondly, the commentaries are selected by the editor before publication, so their quality is guaranteed at least to some extent. Critically, no current PPPR has the features comparable to these in BBS.
(7) Consider revising. While this is true for BBS, I don’t see how a journal of peer reviews that is open to all articles within a field, including preprints, as the target papers could replicate the prestige of BBS. This passage doesn’t seem to help the argument in favor of the new system as currently proposed. However, you might revise the proposal. For example, I could imagine a “Journal of Peer Commentary in Psychology” applying the BBS model to editorially selected papers of broad interest.
To summarize, we might be able to create a new and better PPPR system by simply combining the advantages of BBS commentary – (1) strong incentive for commenters and (2) high readability – with those of the current PPPRs – (3) unlimited target selection and (4) unlimited commentary accumulation -. In the next section, we propose a possible blueprint for the implementation of these ideas, especially with a focus on the first two, because the rest has already been realized in the current media.
(8) Consider revising. The first two points seem at a strong tension with the second two points. Strong incentive to review requires highly visible target publications, which isn’t possible if target selection is unlimited. High readability also appears compromised when reviews come in over a long period and there is no limit to their number. This should at least be discussed.
Among the features that seem critical to the successful implementation of PPPR, strong incentives for commenters is probably the most important factor. We speculated that BBS has achieved this goal by providing the commentaries a status equivalent to a standard academic paper. Furthermore, this is probably realized by the journal’s two unique characteristics: its academic prestige and the selection of commentaries by the editor. Based on these considerations, we propose the following plans for the new PPPR system.
(9) Consider revising. As discussed above, the commentaries do not quite have “equivalent” status to a standard academic paper.

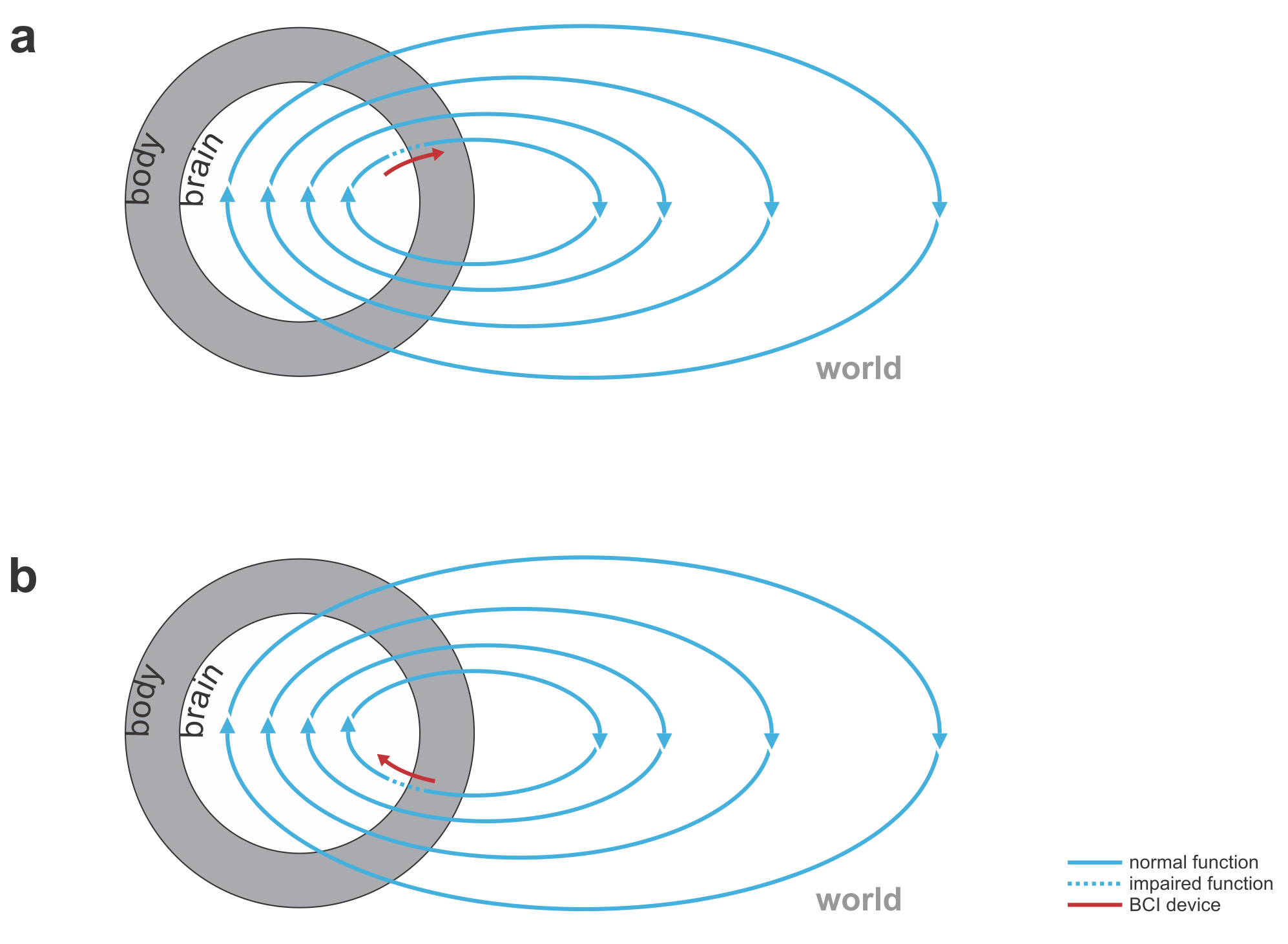
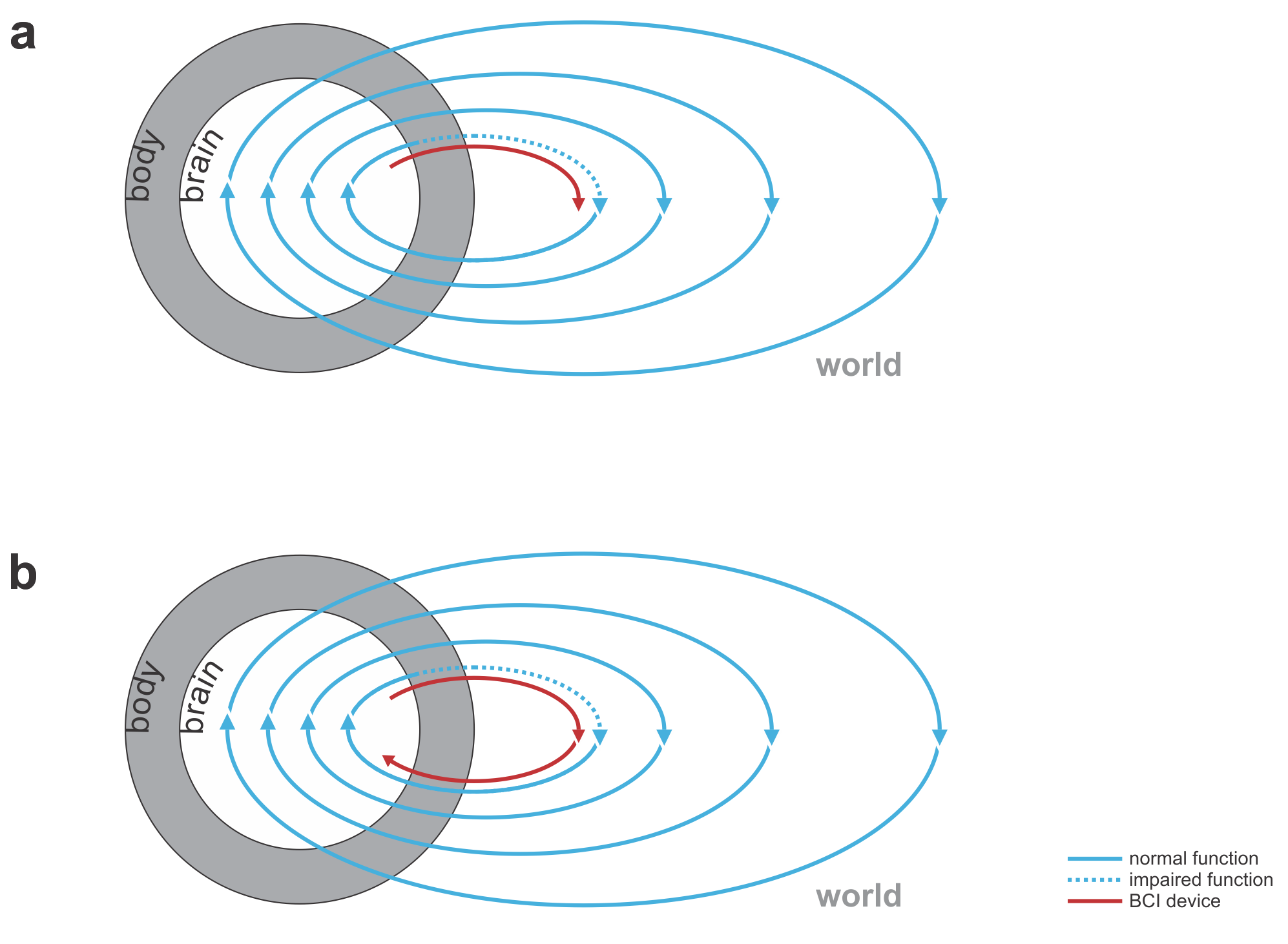 Fig. 1 | Uni- and bidirectional prosthetic-control BCIs. (a) A unidirectional BCI (red) for control of a prosthetic hand that reads out neural signals from motor cortex. The patient controls the hand using visual feedback (blue arrow). (b) A bidirectional BCI (red) for control of a prosthetic hand that reads out neural signals from motor cortex and feeds back tactile sensory signals acquired through artificial sensors to somatosensory cortex.
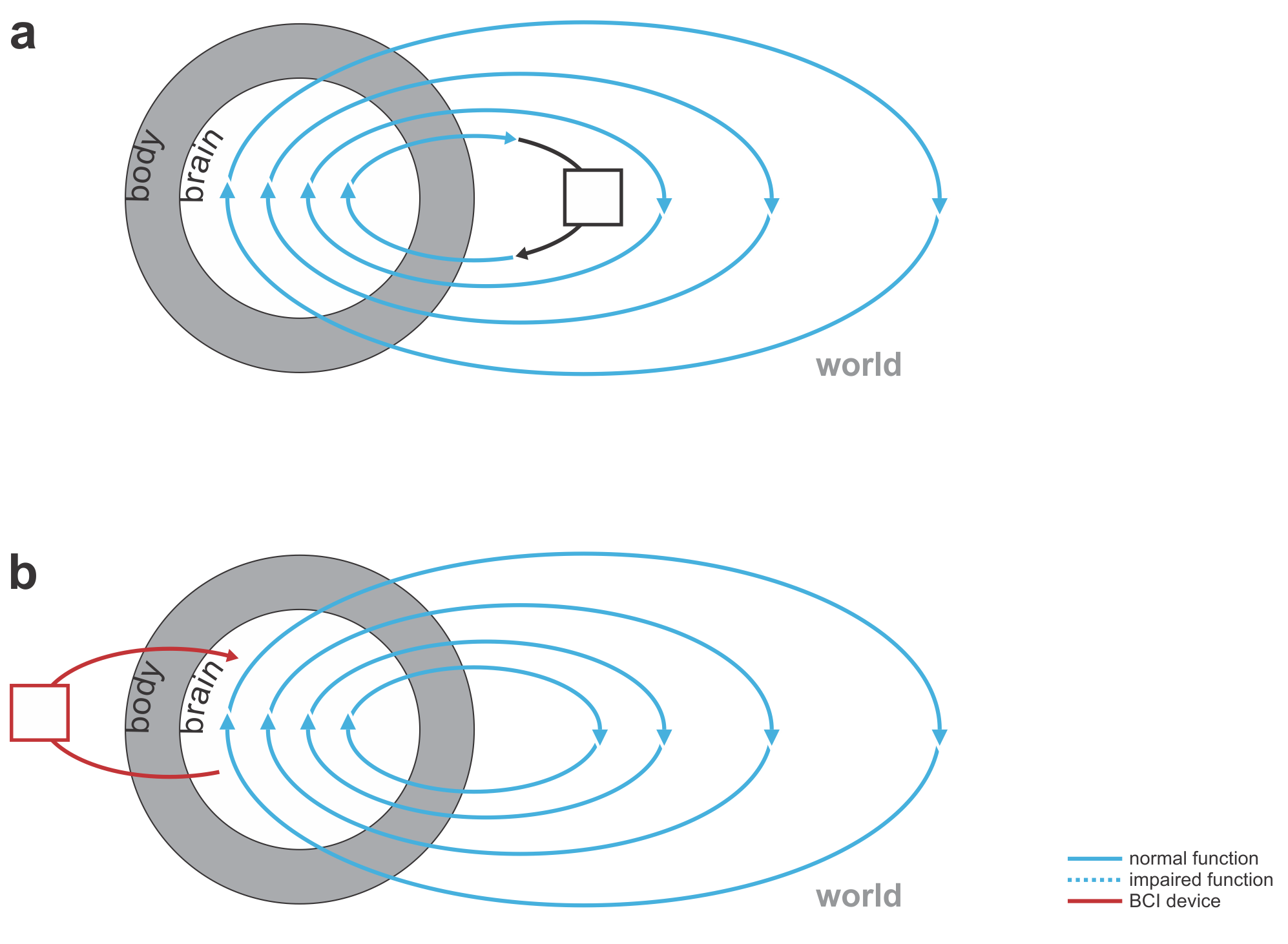
Fig. 1 | Uni- and bidirectional prosthetic-control BCIs. (a) A unidirectional BCI (red) for control of a prosthetic hand that reads out neural signals from motor cortex. The patient controls the hand using visual feedback (blue arrow). (b) A bidirectional BCI (red) for control of a prosthetic hand that reads out neural signals from motor cortex and feeds back tactile sensory signals acquired through artificial sensors to somatosensory cortex.